Cleaning Srm Data In Skyline
First steps, done on 8/8
Clean up data in Skyline:
Used final version of sequence file (which I uploaded from the Vantage computer to Owl @ UWPR on 7/28) to associate .raw files with samples - determine which samples are which for the duplicate named files. The final sequence file, which will also contain the time loaded into tray & injection time (in case this is a factor), is located in the Data folder of this repo, here.

Make sure I have all data in Skyline
Confirmed in Skyline by going to Edit -> Manage Results & cross-checking data file names in Skyline with the sequence file, and the .raw files on Owl.

Confirm correct peaks picked & determine if any transitions/peptides are un-usable
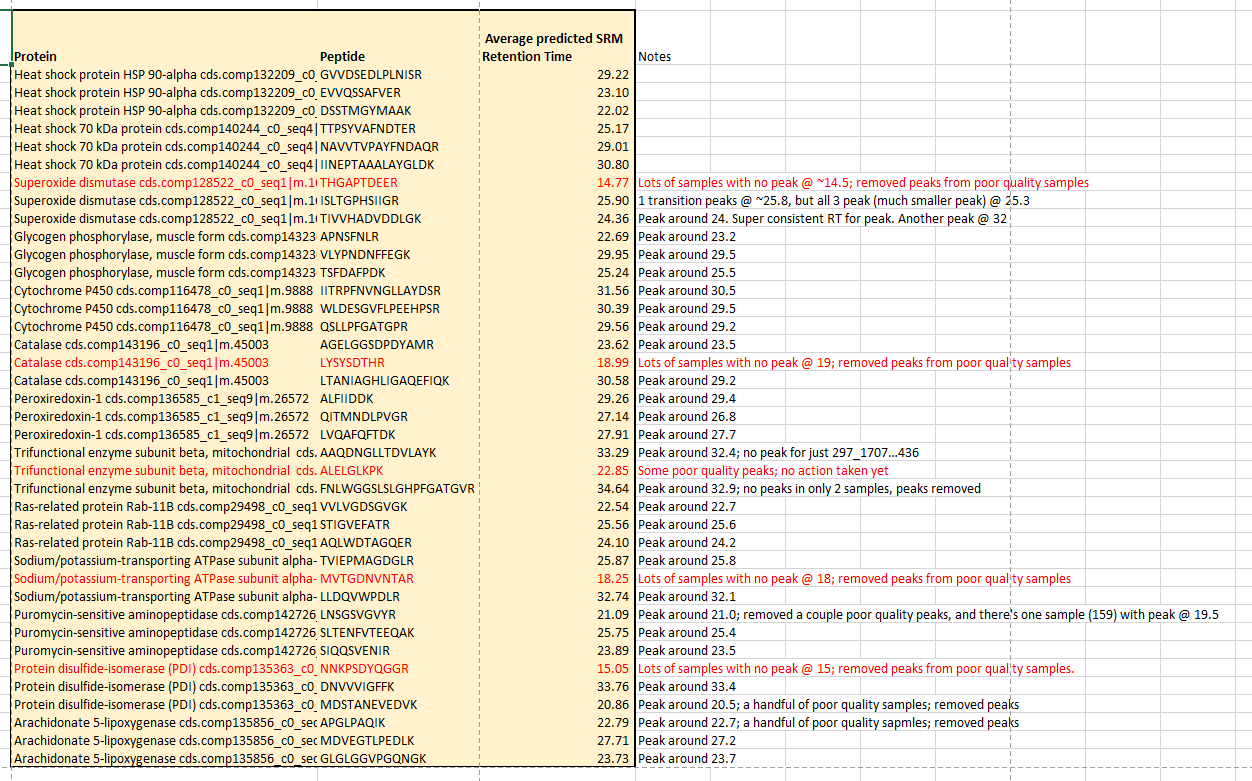
Referenced the Predicted SRM Retention Time calculations I did at the beginning of my mass spec run to confirm correct peaks picked. ID’d peptides with lots of noice, and no clear peak at the predicted RT. Here is a screen shot of the predicted RT, with poor quality peptides in red. NOTE: Need at least 2 transitions for each peptide to move forward with analysis.
Next steps:
- Adjust peak boundaries
- Determine if any .raw files should be discarded, based on poor-quality data and those that I re-ran & re-made
- Look @ all blank runs to see if there are any weird signals
- Check out blank samples
- Download data! Protein, peptide, transition, area, retention time
- Work through Emma’s suggestions: normalization, dilution durve check, NMDS, ROC curves, data upload
