C. magister MiSeq analysis continued
I ran the MiSeq data through the trimming/filtering/alignment pipeline, and a handful of the samples have very low %CpG methylation compared to the others: samples 5, 6, 9, 13, 14, & kind-of 19. Check out the Bismark summary report, and the MultiQC report (on trimmed/filtered reads). In an effort to identify possible causes, I looked at correlations among %CpG meth and various library prep and sequencing metrics (from this table, processed in this RMarkdown notebook). Here’s a big correlation plot with all library prep / sequencing metrics (CpGmeth is 3rd from the bottom/right).

The strongest correlation is with “%total reads that are unique”:

Here’s also %meth ~ coverage plots for each sample

But some other factors also correlate loosely with %CpG meth

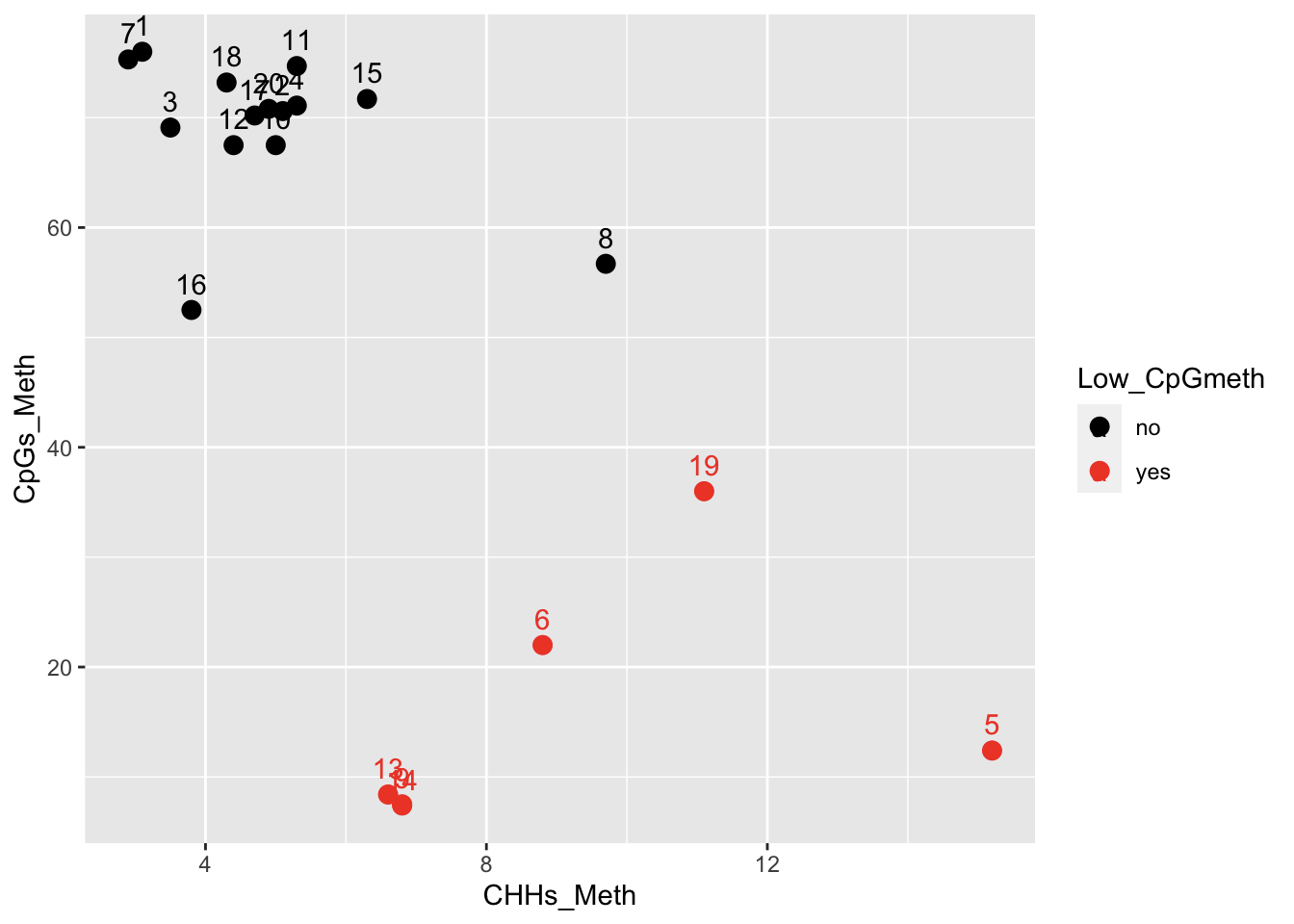
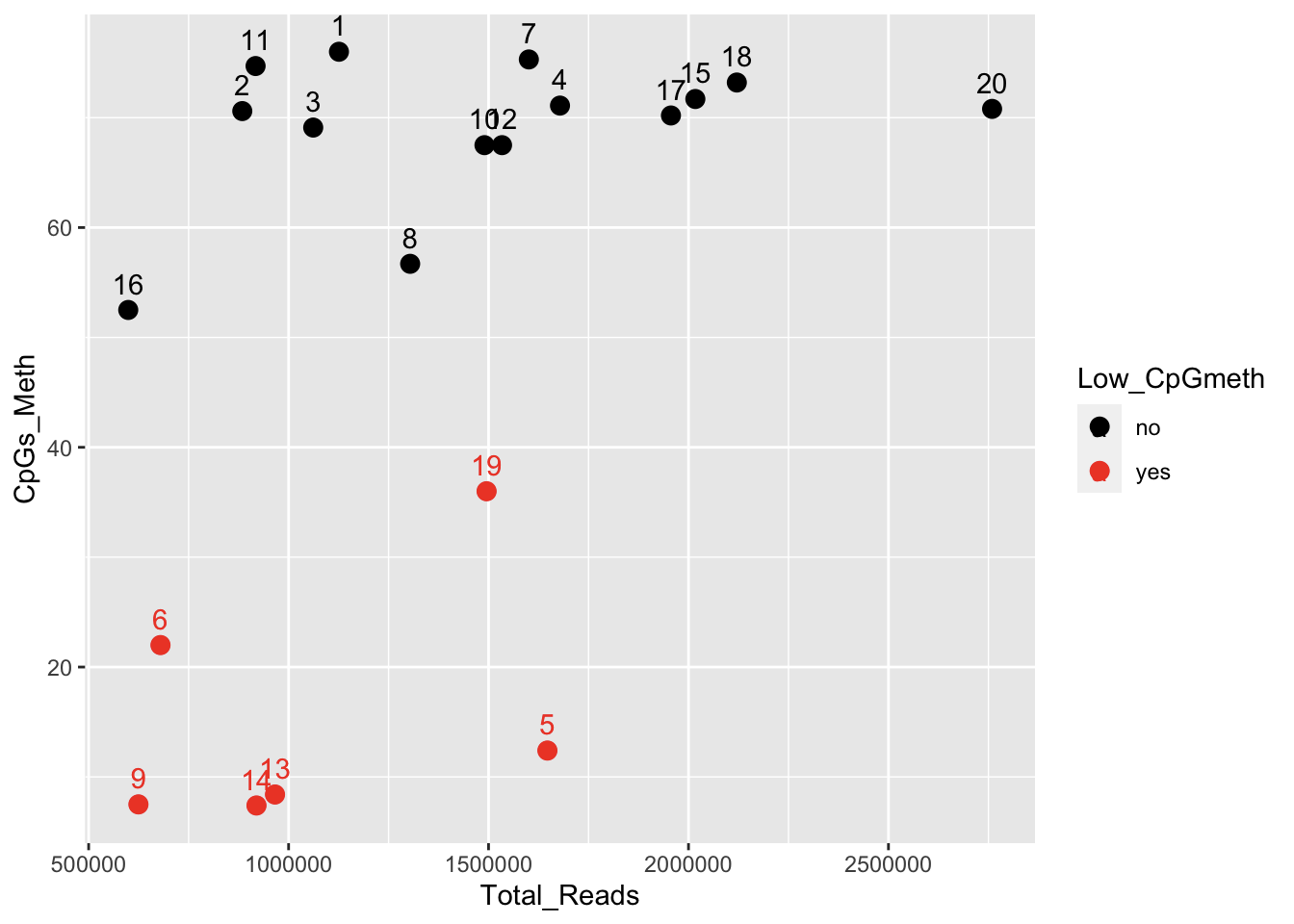

Contacting Sam to see if he has any thoughts regarding library prep, and possible impacts to alignment and %meth.
Written on December 27, 2020
