QuantSeq library amplification, purification, and QA
The final step in the QuantSeq library prep is to amplify my cDNA libraries (using the optimal cycle number) and then purify. Finally, to assess quantity and quality of finished libraries I use the Qubit High Sensitivity DNA kit to measure cDNA concentration, and the Bioanalyzer High Sensitivity DNA chip kit to measure fragment lengths. I worked in batches based on the number of cycles needed to amplify.
IMPORTANT NOTE: I have 2 sets of indexes with the same numbers. I therefore prepped the ctenidia samples with one set, and the larvae with another set. Therefore, I SHOULD NOT run ctenidia and larval samples in the same lane (since two samples have the same index number).
The spreadsheet where I have all this information organized is in the laura-quantseq repo, file is: 2019-July_RNA-isolation-ctenidia-larvae.xlsx
12/30/2019 - 16 cycles
- I noticed that I didn’t have the full 17 uL in any of the ds cDNA libraries, probably missing ~ 2uL. Not sure why, possibly due to pipette loss?
- Sample #331 volume depleted after PCR, maybe due to evaporation b/c there was a small crease in the foil? This was located at well A1 - should ensure tight seal for all future plates.
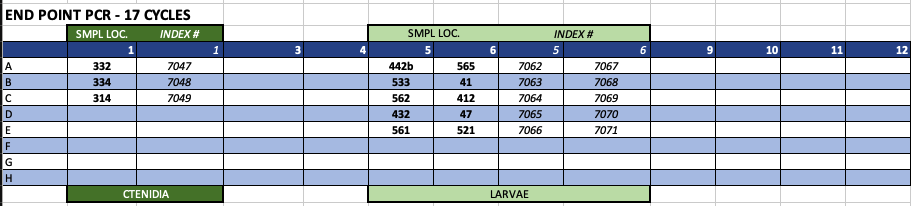
Amplification plate configuration

1/1/2020 - 14 cycles
- Again, samples <17 uL.
Amplification plate configuration

1/2/2020 - 15 cycles
- Again, most samples <17 uL.
Amplification plate configuration

1/4/2020 - 17, 18 and 20 cycles
- I amplified 3 batches of samples today - those needing 17, 18 and 20 cycles (ran in that order). I held amplified samples at 10C until all groups were done, then combined all samples onto one plate, purified, and quantified.
- Unfortunately, none of these libraries amplified! I don’t know whether there was an error in the amplification step, OR if the high number of cycle needed represent low concentration/quality. Looking/thinking back through all my steps I do not see any possible errors that could have been made in the PCR mix (it’ a simple master mix using 2 reagents), or during purification. Regardless, I will need to re-do all these libraries!
Amplification plate configuration & index


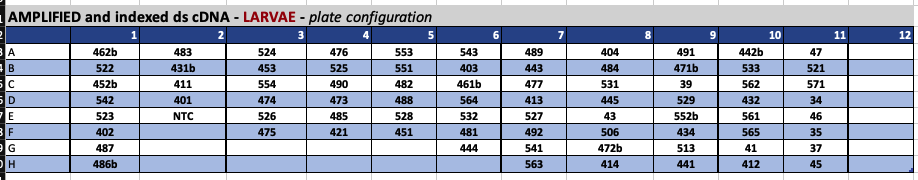
Amplified and Indexed QuantSeq cDNA libraries
This is the plate configuration of the finished libraries. Based on the quantification/Bioanalyzer results, some of these libraries will need to be re-done. However, this is an important map!

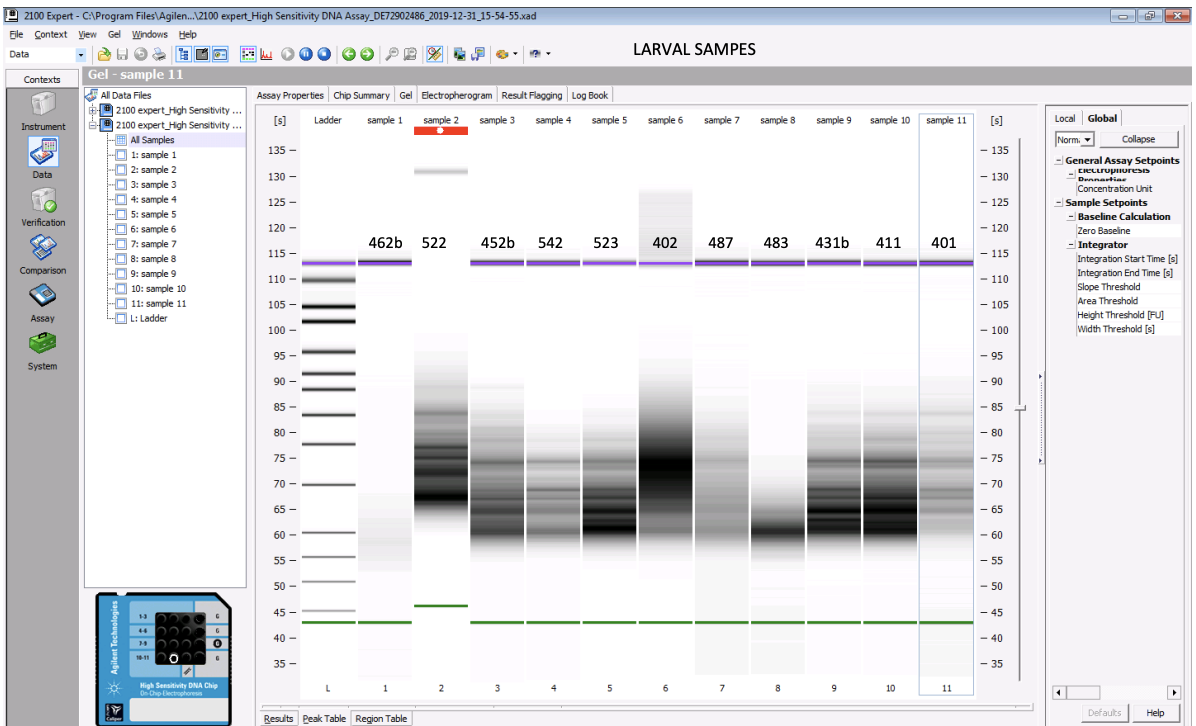
Bioanalyzer quality check
- I ran 1-2 chips per amplification group. The first 2 chips, which I did for the 16 cycle group, worked great! Results below.
- I had some issues with the next few chips. Not sure why, but I suspect that it had to do with the fact that I used the Seeb lab’s pipette/pipette tips.
- I have other chip results, but did not take screen shots at the time - will follow up with those, but for the time being all files are saved in the repo: laura-quantseq/data/library-prep
Results for samples ran for 16 cycles